Detection of Glucose with a Self-Made Biosensor 3/5 – Immobilization of Enzymatic Biosensors
This chapter is part of the series ‘Detection of glucose with a self-made biosensor based on glucose oxidase’. In this section, we elaborate on the workings of biosensors.
Immobilization of Enzymatic Biosensors
Biosensors based on enzymes are divided into direct and indirect electron transfer systems. A direct electron transfer means that the electrons are directly transferred from or to the active center of the enzyme to the electrode. An indirect electron transfer uses a redox mediator or the product of the enzymatic reaction to transport electrons between the active center of the enzyme and the electrode. Direct electron transfer is more efficient once it has been established, but immobilizing an enzyme for direct electron transfer is quite difficult. The active center is inside an insulating protein shell. Above a certain distance electron transfer becomes impossible. The conditions are described by the Marcus theory [1]. Whether a direct electron transfer is possible depends on the enzyme structure, that is if the enzyme does have a thick protein shell around the active center, direct electron transfer isn’t possible.
Adsorption
It is difficult to say whether the large number of immobilization methods present today has been developed despite or because of these challenges. An easy method for immobilizing biological recognition elements is plain adsorption (see Figure 2.1).
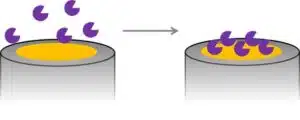
Some electrode materials interact with proteins strongly enough so that they will not be washed away. For example, many carbon species show π-π as well as van-der-Waal’s interaction and often a mixture of functional groups. Gold surfaces bind sulfur-containing amino acids like cysteine. Although this technique is very easy to perform and thus popular especially in the first research steps, the adsorbed proteins are sensitive to changes of ionic strength, that is the number of ions in solution, the pH, as well as temperature, and can thus desorb from the surface if a condition changes. The direct contact with an electrode surface is very likely to cause denaturation of the adsorbed protein. It is possible to modify the electrode by covering it with a monolayer that creates a suitable environment for the enzyme, that does not cause denaturation, and that allows electron transfer (mediated or directly). An example of such a surface modification is a self-assembled monolayer of pyridine thiol on a gold electrode. If cytochrome c is adsorbed to that surface, the electron transfer between cytochrome c and the electrode is improved compared to a blank electrode.
Covalent binding
Another technique is the covalent binding of the proteins to the electrode. Self-assembled monolayers, for example, can provide the necessary functional groups to create a covalent bonding (see Figure 2.2). A 3-sulfanylpropanoic acid layer on a gold surface will bind to the gold surface with the sulfur group and form a self-assembled monolayer. The carboxylic groups can be activated and will form an ester bound with the amine groups of biological recognition elements and will thus bind the biological recognition element to the surface.
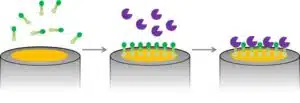
A covalent bond is stable and the immobilization is not sensitive anymore to the conditions of the solution surrounding the electrode. If the position of the functional group at the recognition element that is used for the covalent bond is known, the architecture of the surface after the immobilization is known and defined as well. Activating the electrode surface for creating a suitable functional group for the covalent bond is often the difficult part.
Adsorption and covalent bonding are suitable for mediated and direct electron transfer biosensors. A mediated transfer needs either the products of the enzymatic reaction to be the mediator or a co-immobilized redox mediator on the electrode surface. If no mediator is immobilized, a free diffusing redox mediator needs to be added to the sample solution.
Entrapment in a polymer matrix
A very different approach is the entrapment of an enzyme in a polymer matrix. The idea is to create a layer of polymer on the electrode surface that has enzymes captured inside like a fisher net. The enzyme can only work if the polymer does not interfere with the enzymatic reaction. If the polymer deforms the enzyme, blocks the diffusion of the analyte to the enzyme, the diffusion of the enzyme’s products away from the enzyme, or the movement of the mediator between the enzyme and the electrode, the biosensor won’t work. Since polymers are produced with a huge variety of structures and properties a lot of polymer-based biosensors are known nowadays.
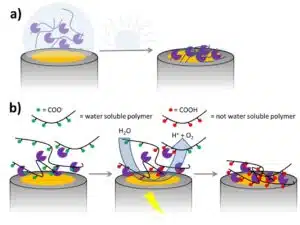
The polymer and enzyme matrix can be applied to the electrode in different ways. A very simple approach is drop coating. The polymer and enzyme are dissolved in an organic solvent. A drop of this solution is positioned on the working electrode and dried (see Figure 2.3a). Another way is to use a polymer that is only soluble in an aqueous solution in a certain pH range.
After mixing polymer and enzyme solution, the electrode is dipped into the polymer solution and then in a solution outside the pH range of the polymer. This is repeated a number of times. Another option is for the electrode to be immersed in the polymer solution and the pH is changed only locally by applying a high cathodic or anodic potential (see Figure 2.3b). The polymer will precipitate in the vicinity of the electrode.
Electropolymerization
Another approach is creating bonds to form the polymer in the presence of the enzyme. The molecules that will form the polymer are called monomers. There are different ways to perform the polymerization. A common way is to add a cross-linker solution, in which the monomers have already formed short chains (oligomers) that are still soluble or at least form a suspension. The cross-linkers form bonds between the oligomers and they precipitate as a polymer. Another elegant way is to perform an electropolymerization. The monomer is activated by applying a potential to the electrode. The activation is usually the formation of a radical that attacks another monomer resulting in a dimer radical. The process is repeated until the polymer is too long to stay in solution and precipitates (see Figure 2.4).
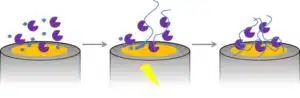
A common example of this technique is the formation of polypyrrole. The advantage of pyrrole is that the monomer is more difficult to oxidize than its dimers, trimers, etc., so that a chain length growth is more probable than the start of a new chain [2]. If a radical reaction is used, as in the case of polypyrrole, the polymerization can be improved by removing oxygen from the monomer solution, because oxygen is a radical scavenger, due to its nature as a diradical.
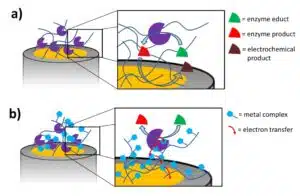
Furthermore, polypyrrole has the advantage of being a conducting polymer. The growth is not limited and can be controlled by the applied potential or current and the time it is applied. The electrons can also be easily transferred to the electrode. While non-conducting polymers require a mediator or an electrochemical active product of the enzymatic reaction to travel to the electrode (see Figure 5a), a conducting polymer can convert the mediator or product close to the enzyme and transport the electrons. In an ideal case, the polymer is able to collect the electrons directly from the active center of the enzyme. In this case the redox mediator also does not have to be added to the solution. Polymers modified with metal complexes, for instance osmium, have shown to be able to do this. The polymer contains an osmium complex, whose shell allows the complex to diffuse into the active center of the enzyme and accept or donate electrons. The electrons will hop from complex to complex till they reach the electrode (see Figure 2.5 b). Of course, the distance between the complexes and their flexibility is chosen in such a way that an electron transfer between the complexes is possible.
Literature
[1] Borgmann, S.; Hartwich, G.; Schulte, S.; Schuhmann, W.; Amperometric Enzyme Sensors based on Direct and Mediated Electron Transfer, Perspectives in Bioanalysis, 2005 (1), p. 599.
[2] Schuhmann, W.; Conducting Polymer Based Amperometric Enzyme Electrodes; Mikrochim. Acta, 1995 (121); p. 1
